General information
| Regulator | AbrC3 |
|---|---|
| Gene identifier | SCO4596 - SCD20.14c |
| Family | NarL |
| Regulation type | Activator |
| Function | Positive response regulator of antibiotic production |
| Organism | Streptomyces coelicolor |
Sequence logo
Logo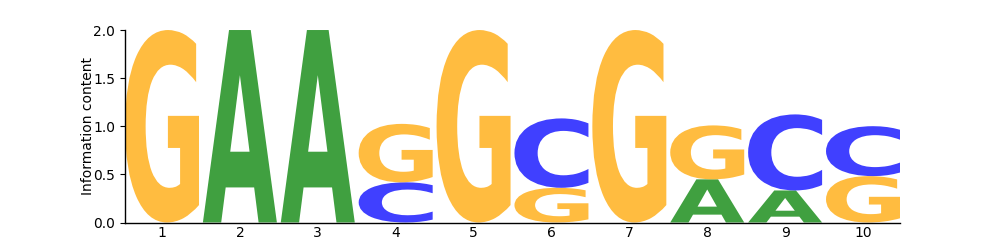
Based on 27 sites.
Position Frequency Matrix
PFM| T | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|---|---|---|---|---|---|---|---|---|---|---|
| G | 27 | 0 | 0 | 16 | 27 | 9 | 27 | 15 | 0 | 13 |
| C | 0 | 0 | 0 | 11 | 0 | 18 | 0 | 0 | 19 | 14 |
| A | 0 | 27 | 27 | 0 | 0 | 0 | 0 | 12 | 8 | 0 |
Position Weight Matrix
PWM| T | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 |
|---|---|---|---|---|---|---|---|---|---|---|
| G | 2.14 | -1.32 | -1.32 | 1.47 | 2.14 | 0.79 | 2.14 | 1.39 | -1.32 | 1.22 |
| C | -1.32 | -1.32 | -1.32 | 1.02 | -1.32 | 1.62 | -1.32 | -1.32 | 1.68 | 1.31 |
| A | -1.32 | 2.14 | 2.14 | -1.32 | -1.32 | -1.32 | -1.32 | 1.12 | 0.66 | -1.32 |
| Animate |
|
| Node Repulsion | |
| Ideal Edge Length | |
Loading...
| Gene ID | Location | Orientation | Binding sequence | Method | Reference |
|---|---|---|---|---|---|
| SCO0167 | 354 | + | GAAGGGGGCG | ChIP-chip | S. Rico et al. (2014) |
| SCO0951 | -329 | - | GGTCGCCTTC | ChIP-chip | S. Rico et al. (2014) |
| SCO1391 | 1113 | + | GAAGGCGGCG | ChIP-chip | S. Rico et al. (2014) |
| SCO3263 | -240 | - | GTTCGCCTTC | ChIP-chip | S. Rico et al. (2014) |
| SCO3334 | -387 | - | GGCCGCGTTC | ChIP-chip | S. Rico et al. (2014) |
| SCO4123 | -674 | + | GAACGCGACC | ChIP-chip | S. Rico et al. (2014) |
| SCO4596 | -137 | - | CTTCCCGTTC | ChIP-chip | S. Rico et al. (2014) |
| SCO5085 | 72 | + | GAAGGCGACC | ChIP-chip | S. Rico et al. (2014) |
| SCO5236 | -213 | - | GGTCGCGTTC | ChIP-chip | S. Rico et al. (2014) |
| SCO5330 | -975 | + | GAAGGGGGCG | ChIP-chip | S. Rico et al. (2014) |
| SCO5332 | 1134 | - | CTTCGCCTTC | ChIP-chip | S. Rico et al. (2014) |
| SCO5632 | -1261 | + | GAACGGGACG | ChIP-chip | S. Rico et al. (2014) |
| SCO5691 | -777 | + | GAACGCGGCG | ChIP-chip | S. Rico et al. (2014) |
| SCO5912 | -54 | - | CTCCCCCTTC | ChIP-chip | S. Rico et al. (2014) |
| SCO5966 | -803 | + | GAAGGCGGAC | ChIP-chip | S. Rico et al. (2014) |
| SCO6273 | 3138 | - | CGCCCCGTTC | ChIP-chip | S. Rico et al. (2014) |
| SCO6732 | 1140 | + | GAAGGCGGCC | ChIP-chip | S. Rico et al. (2014) |
| SCO6820 | -519 | + | GAACGCGGCC | ChIP-chip | S. Rico et al. (2014) |
| SCO6941 | -306 | - | GGCCGCCTTC | ChIP-chip | S. Rico et al. (2014) |
| SCO6951 | -411 | - | CGCCGCGTTC | ChIP-chip | S. Rico et al. (2014) |
| SCO6992 | -321 | + | GAAGGGGACG | ChIP-chip | S. Rico et al. (2014) |
| SCO7077 | -294 | + | GAAGGCGGCG | ChIP-chip | S. Rico et al. (2014) |
| SCO7545 | -369 | - | GGCCGCGTTC | ChIP-chip | S. Rico et al. (2014) |
| SCO0736 | -53 | + | GAAGGCGACC | ChIP-chip | S. Rico et al. (2014) |
| SCO2113 | -26 | - | GTTCCCGTTC | ChIP-chip | S. Rico et al. (2014) |
| SCO3328 | -46 | + | GAAGGGGAAG | ChIP-chip | S. Rico et al. (2014) |
| SCO6809 | -112 | + | GAAGGCGGAC | ChIP-chip | S. Rico et al. (2014) |
| Region | Location | Orientation | Score | Binding sequence | Method |
|---|

