General information
| Regulator | ArgR |
|---|---|
| Gene identifier | SCO1576 - SCL24.12c |
| Family | ArgR |
| Regulation type | Repressor |
| Function | Regulator of arginine biosynthesis genes |
| Organism | Streptomyces coelicolor |
Sequence logo
Logo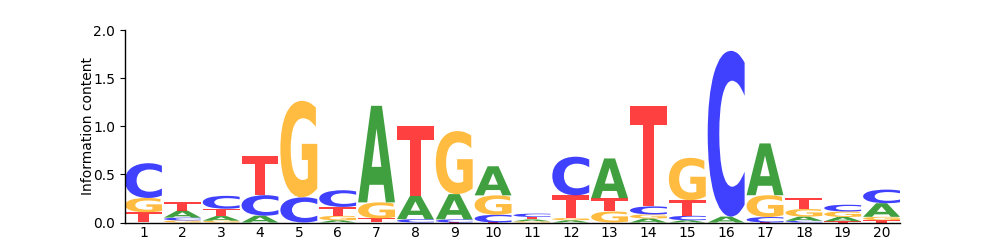
Based on 29 sites.
Position Frequency Matrix
PFM| T | 5 | 12 | 8 | 17 | 0 | 8 | 1 | 21 | 0 | 0 | 8 | 10 | 6 | 25 | 7 | 0 | 0 | 13 | 2 | 2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | 7 | 3 | 2 | 0 | 23 | 4 | 4 | 0 | 20 | 10 | 4 | 1 | 5 | 1 | 19 | 0 | 8 | 9 | 9 | 3 |
| C | 17 | 4 | 14 | 9 | 6 | 15 | 0 | 1 | 1 | 4 | 11 | 17 | 0 | 2 | 2 | 28 | 2 | 2 | 11 | 12 |
| A | 0 | 10 | 5 | 3 | 0 | 2 | 24 | 7 | 8 | 15 | 6 | 1 | 18 | 1 | 1 | 1 | 19 | 5 | 7 | 12 |
Position Weight Matrix
PWM| T | 0.12 | 1.04 | 0.59 | 1.46 | -1.32 | 0.59 | -0.89 | 1.72 | -1.32 | -1.32 | 0.59 | 0.83 | 0.30 | 1.94 | 0.45 | -1.32 | -1.32 | 1.13 | -0.57 | -0.57 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | 0.45 | -0.30 | -0.57 | -1.32 | 1.84 | -0.07 | -0.07 | -1.32 | 1.66 | 0.83 | -0.07 | -0.89 | 0.12 | -0.89 | 1.59 | -1.32 | 0.59 | 0.71 | 0.71 | -0.30 |
| C | 1.46 | -0.07 | 1.22 | 0.71 | 0.30 | 1.30 | -1.32 | -0.89 | -0.89 | -0.07 | 0.94 | 1.46 | -1.32 | -0.57 | -0.57 | 2.09 | -0.57 | -0.57 | 0.94 | 1.04 |
| A | -1.32 | 0.83 | 0.12 | -0.30 | -1.32 | -0.57 | 1.89 | 0.45 | 0.59 | 1.30 | 0.30 | -0.89 | 1.53 | -0.89 | -0.89 | -0.89 | 1.59 | 0.12 | 0.45 | 1.04 |
Hidden Markov Model (HMM)
| Consensus | TG-ATG-2-CAT-CA |
|---|---|
| Alignment length | 20 |
| Animate |
|
| Node Repulsion | |
| Ideal Edge Length | |
Loading...
| Gene ID | Location | Orientation | Binding sequence | Method | Reference |
|---|---|---|---|---|---|
| SCO1086 | -24 | + | GAACGCATACCTATGCAGTG | DNAse footprinting | R. Pérez-Redondo et al. (2012) |
| SCO2015 | -158 | + | CCACGCATGACCACTCAACA | DNAse footprinting | R. Pérez-Redondo et al. (2012) |
| SCO2686 | -22 | + | CAATGCATGATCATGCCACA | DNAse footprinting | R. Pérez-Redondo et al. (2012) |
| SCO3034 | -139 | + | GGAACCAAGCCCATGCAGTA | DNAse footprinting | R. Pérez-Redondo et al. (2012) |
| SCO3979 | -34 | + | TAACGGATAGCTTTTCATAA | DNAse footprinting | R. Pérez-Redondo et al. (2012) |
| SCO4293 | 8 | + | GCCTCCATGGCTGTGCAGAC | DNAse footprinting | R. Pérez-Redondo et al. (2012) |
| SCO5583 | -234 | + | CCATGCCAGGTCATTCGGAG | DNAse footprinting | R. Pérez-Redondo et al. (2012) |
| SCO1086 | -4 | + | CAGTCAATGACTTTTCAGGT | DNAse footprinting | A. Botas et al. (2018) |
| SCO1487 | -8 | + | CTCCGTAAGGCGATTCATGC | DNAse footprinting | A. Botas et al. (2018) |
| SCO1488 | 14 | + | TGTTGCTTGTCCATACGAAA | DNAse footprinting | A. Botas et al. (2018) |
| SCO1489 | -62 | + | CGCTGCGTAACCTCACAGTG | DNAse footprinting | A. Botas et al. (2018) |
| SCO1570 | -29 | + | TCATGCATGAGTATGCAGAA | DNAse footprinting | A. Botas et al. (2018) |
| SCO1580 | -17 | + | CGATGCACGTTTATGCAATG | DNAse footprinting | A. Botas et al. (2018) |
| SCO1580 | -37 | + | GAGAGCATGACTATACGTGC | DNAse footprinting | A. Botas et al. (2018) |
| SCO2055 | -62 | + | GTTCGGACGGTCACGCAGTG | DNAse footprinting | A. Botas et al. (2018) |
| SCO2055 | -41 | + | GCATGAACGGTTGTACGAAG | DNAse footprinting | A. Botas et al. (2018) |
| SCO2232 | -236 | + | TGCTGCAAAAATGTGCAAGA | DNAse footprinting | A. Botas et al. (2018) |
| SCO2232 | -124 | + | CTCTGCAAGCTCTTGCCGCC | DNAse footprinting | A. Botas et al. (2018) |
| SCO2529 | -143 | + | GTCCGTATCAGTATGCGGGA | DNAse footprinting | A. Botas et al. (2018) |
| SCO3943 | -38 | + | GGACGCATATGCACGCGTTG | DNAse footprinting | A. Botas et al. (2018) |
| SCO3943 | -68 | + | TTCTGCAAGATCATTAATGC | DNAse footprinting | A. Botas et al. (2018) |
| SCO4158 | -47 | + | CCTTGGATGACCTTGCGCCC | DNAse footprinting | A. Botas et al. (2018) |
| SCO5327 | 8 | + | GCCTCGTTGGTCATGCATCC | DNAse footprinting | A. Botas et al. (2018) |
| SCO5864 | -266 | + | CTCTCCGTGATCATGCACCC | DNAse footprinting | A. Botas et al. (2018) |
| SCO5976 | -34 | + | GTTCGTATAGACTTCCAGAA | DNAse footprinting | A. Botas et al. (2018) |
| SCO5976 | -54 | + | CGCTGTATAGAAATTCAGAA | DNAse footprinting | A. Botas et al. (2018) |
| SCO7036 | 3 | + | TGATGCATACTCTTCCTATG | DNAse footprinting | A. Botas et al. (2018) |
| SCO7036 | -17 | + | CTTTGCATGGTCATGCGTAA | DNAse footprinting | A. Botas et al. (2018) |
| SCO7314 | 14 | + | ATCCGCATGCTCATAGAAAC | DNAse footprinting | A. Botas et al. (2018) |
| Region | Location | Orientation | Score | Binding sequence | Method |
|---|

