General information
| Regulator | CelR |
|---|---|
| Gene identifier | SCO1376 - SC10A9.18 |
| Family | LacI |
| Regulation type | Repressor |
| Function | Cellobiose uptake repressor |
| Organism | Streptomyces coelicolor |
Sequence logo
Logo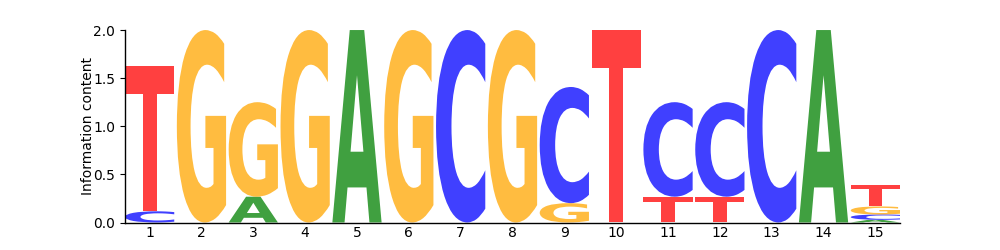
Based on 14 sites.
Position Frequency Matrix
PFM| T | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 3 | 3 | 0 | 0 | 8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | 0 | 14 | 11 | 14 | 0 | 14 | 0 | 14 | 2 | 0 | 0 | 0 | 0 | 0 | 3 |
| C | 1 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 12 | 0 | 11 | 11 | 14 | 0 | 2 |
| A | 0 | 0 | 3 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 1 |
Position Weight Matrix
PWM| T | 2.04 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | 2.14 | 0.33 | 0.33 | -1.32 | -1.32 | 1.43 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | -1.32 | 2.14 | 1.82 | 2.14 | -1.32 | 2.14 | -1.32 | 2.14 | -0.04 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | 0.33 |
| C | -0.54 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | 2.14 | -1.32 | 1.94 | -1.32 | 1.82 | 1.82 | 2.14 | -1.32 | -0.04 |
| A | -1.32 | -1.32 | 0.33 | -1.32 | 2.14 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | 2.14 | -0.54 |
| Animate |
|
| Node Repulsion | |
| Ideal Edge Length | |
Loading...
| Gene ID | Location | Orientation | Binding sequence | Method | Reference |
|---|---|---|---|---|---|
| SCO0643 | -54 | - | CCTTGGGAGCGCTCCCATGC | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO0716 | -33 | + | GCGTGAGAGCGCTCTCAGGC | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO0765 | -34 | - | ATTTGGGAGCGCTCCCATTC | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO1187 | -67 | + | AAGTGGGAGCGCTCCCATCA | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO1187 | -41 | + | AGTTGGGAGCGCTCCCGCAC | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO2794 | -218 | - | GTGTGGGAGCGCTCCCACAA | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO2795 | -127 | + | TTGTGGGAGCGCTCCCACAC | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO2798 | 4 | + | CTATGGGAGCGCTTCCATGC | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO6546 | -59 | - | CTATGGGAGCGCTCCCACTG | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO6546 | -201 | - | TGATGGAACCGCTCCCACTG | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO6548 | -298 | + | CAGTGGGAGCGGTTCCATCA | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO6665 | -65 | - | CTCTGAGAGCGCTCTCAGAA | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO7559 | -42 | - | CGCTGAGAGCGCTCTCAGAA | DNAse footprinting | P. Tocquin et al. (2016) |
| SCO7637 | 29 | - | CTTTGGGAGCGCTCCCATCG | DNAse footprinting | P. Tocquin et al. (2016) |
| Region | Location | Orientation | Score | Binding sequence | Method |
|---|

