General information
| Regulator | NdgR |
|---|---|
| Gene identifier | SCO5552 - SC1C2.33c |
| Family | IclR |
| Regulation type | Activator |
| Function | Isopropylmalate dehydratase |
| Organism | Streptomyces coelicolor |
Sequence logo
Logo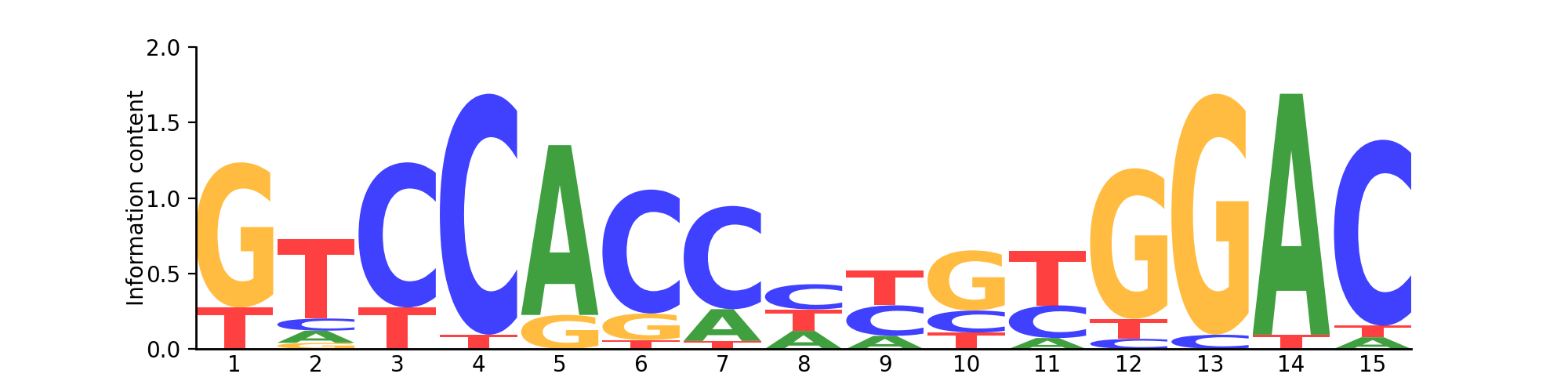
Based on 18 sites.
Position Frequency Matrix
PFM| T | 4 | 13 | 4 | 1 | 0 | 1 | 1 | 6 | 8 | 3 | 10 | 2 | 0 | 1 | 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | 14 | 1 | 0 | 0 | 3 | 3 | 0 | 0 | 0 | 11 | 0 | 15 | 17 | 0 | 0 |
| C | 0 | 2 | 14 | 17 | 0 | 14 | 13 | 7 | 7 | 4 | 6 | 1 | 1 | 0 | 16 |
| A | 0 | 2 | 0 | 0 | 15 | 0 | 4 | 5 | 3 | 0 | 2 | 0 | 0 | 17 | 1 |
Position Weight Matrix
PWM| T | 0.37 | 1.72 | 0.37 | -0.68 | -1.32 | -0.68 | -0.68 | 0.79 | 1.12 | 0.09 | 1.39 | -0.24 | -1.32 | -0.68 | -0.68 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | 1.81 | -0.68 | -1.32 | -1.32 | 0.09 | 0.09 | -1.32 | -1.32 | -1.32 | 1.51 | -1.32 | 1.90 | 2.06 | -1.32 | -1.32 |
| C | -1.32 | -0.24 | 1.81 | 2.06 | -1.32 | 1.81 | 1.72 | 0.97 | 0.97 | 0.37 | 0.79 | -0.68 | -0.68 | -1.32 | 1.98 |
| A | -1.32 | -0.24 | -1.32 | -1.32 | 1.90 | -1.32 | 0.37 | 0.60 | 0.09 | -1.32 | -0.24 | -1.32 | -1.32 | 2.06 | -0.68 |
Hidden Markov Model (HMM)
| Consensus | GTCCACC-2-GTGGAC |
|---|---|
| Alignment length | 15 |
| Animate |
|
| Node Repulsion | |
| Ideal Edge Length | |
Loading...
| Gene ID | Location | Orientation | Binding sequence | Method | Reference |
|---|---|---|---|---|---|
| SCO1553 | 1662781:1662795 | - | GTCCACCCACCGGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO1707 | 1828423:1828437 | + | GTCCACCACGCGGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO1776 | 1897508:1897522 | + | GTCCATCCTGCGGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO2103 | 2261989:2262003 | + | GTCCACCCTTTGGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO2912 | 3163207:3163221 | - | GATCACACTCCGGAA | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO2999 | 3269922:3269936 | + | GTTCACCTCGTGGTC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO3346 | 3704846:3704860 | + | TTCCACCTTGATCAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO4164 | 4580694:4580708 | + | TCCCACTCCTTGGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO4193 | 4601788:4601802 | - | GGTCAGCTCCTGGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO5277 | 5742420:5742434 | + | GACCACCTCGTGGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO5394 | 5863483:5863497 | + | GTCCACACCGTGGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO5412 | 5882573:5882587 | - | GTCCGCCTTGAGGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO5511 | 6002862:6002876 | + | TCCCACCCACTTGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO5522 | 6015204:6015218 | - | GTCCGCCATGCGGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO5552 | 6051783:6051797 | + | GTCCAGAACGCCGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO5561 | 6059774:6059788 | - | GTCCAGCAAGTGGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO6102 | 6702861:6702875 | - | GTCCACATTTTGGAT | ChIP-Seq | J.N. Kim et al. (2015) |
| SCO6510 | 7200390:7200404 | + | TTTTGCCATGTGGAC | ChIP-Seq | J.N. Kim et al. (2015) |
| Region | Location | Orientation | Score | Binding sequence | Method |
|---|

