General information
| Regulator | OsdR |
|---|---|
| Gene identifier | SCO0204 - SCJ12.16c |
| Family | LuxR |
| Regulation type | Activator/repressor |
| Function | Development and stress management regulator |
| Organism | Streptomyces coelicolor |
Sequence logo
Logo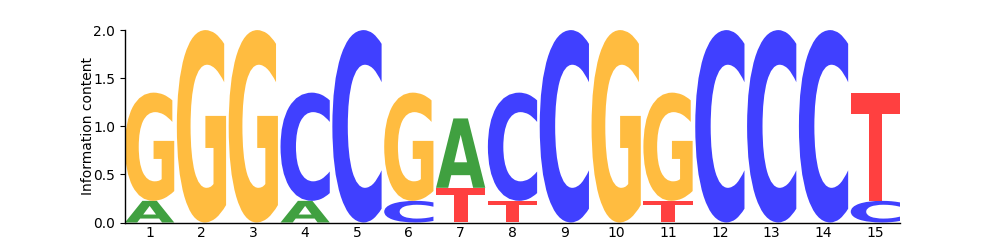
Based on 6 sites.
Position Frequency Matrix
PFM| T | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | 5 | 6 | 6 | 0 | 0 | 5 | 0 | 0 | 0 | 6 | 5 | 0 | 0 | 0 | 0 |
| C | 0 | 0 | 0 | 5 | 6 | 1 | 0 | 5 | 6 | 0 | 0 | 6 | 6 | 6 | 1 |
| A | 1 | 0 | 0 | 1 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Position Weight Matrix
PWM| T | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | 0.79 | 0.09 | -1.32 | -1.32 | 0.09 | -1.32 | -1.32 | -1.32 | 1.90 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | 1.90 | 2.14 | 2.14 | -1.32 | -1.32 | 1.90 | -1.32 | -1.32 | -1.32 | 2.14 | 1.90 | -1.32 | -1.32 | -1.32 | -1.32 |
| C | -1.32 | -1.32 | -1.32 | 1.90 | 2.14 | 0.09 | -1.32 | 1.90 | 2.14 | -1.32 | -1.32 | 2.14 | 2.14 | 2.14 | 0.09 |
| A | 0.09 | -1.32 | -1.32 | 0.09 | -1.32 | -1.32 | 1.62 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 | -1.32 |
| Animate |
|
| Node Repulsion | |
| Ideal Edge Length | |
Loading...
| Gene ID | Location | Orientation | Binding sequence | Method | Reference |
|---|---|---|---|---|---|
| SCO0204 | -64 | + | AGGGCCGGTCGGCCCC | EMSA | M. Urem et al. (2016) |
| SCO0200 | -83 | + | GGGGCCGACCGTCCCT | EMSA | M. Urem et al. (2016) |
| SCO0208 | -86 | + | CGGGCCGACCGGCCCT | EMSA | M. Urem et al. (2016) |
| SCO5978 | -51 | + | CGGGACCTTCGGCCCT | EMSA | M. Urem et al. (2016) |
| SCO2637 | -37 | + | AGGGCCGGTCGGCCTT | EMSA | M. Urem et al. (2016) |
| SCO6041 | -35 | + | GGGGCCGTCCGGCCCC | EMSA | M. Urem et al. (2016) |
| Region | Location | Orientation | Score | Binding sequence | Method |
|---|

